Computational Research
We use bioinformatics to investigate questions in molecular evolution. Tree building animation
Tree building animation
Bioinformatics is the field of science at the intersection of biology and computer science, where computational approaches are applied to the analysis of complex biological data.
Our research
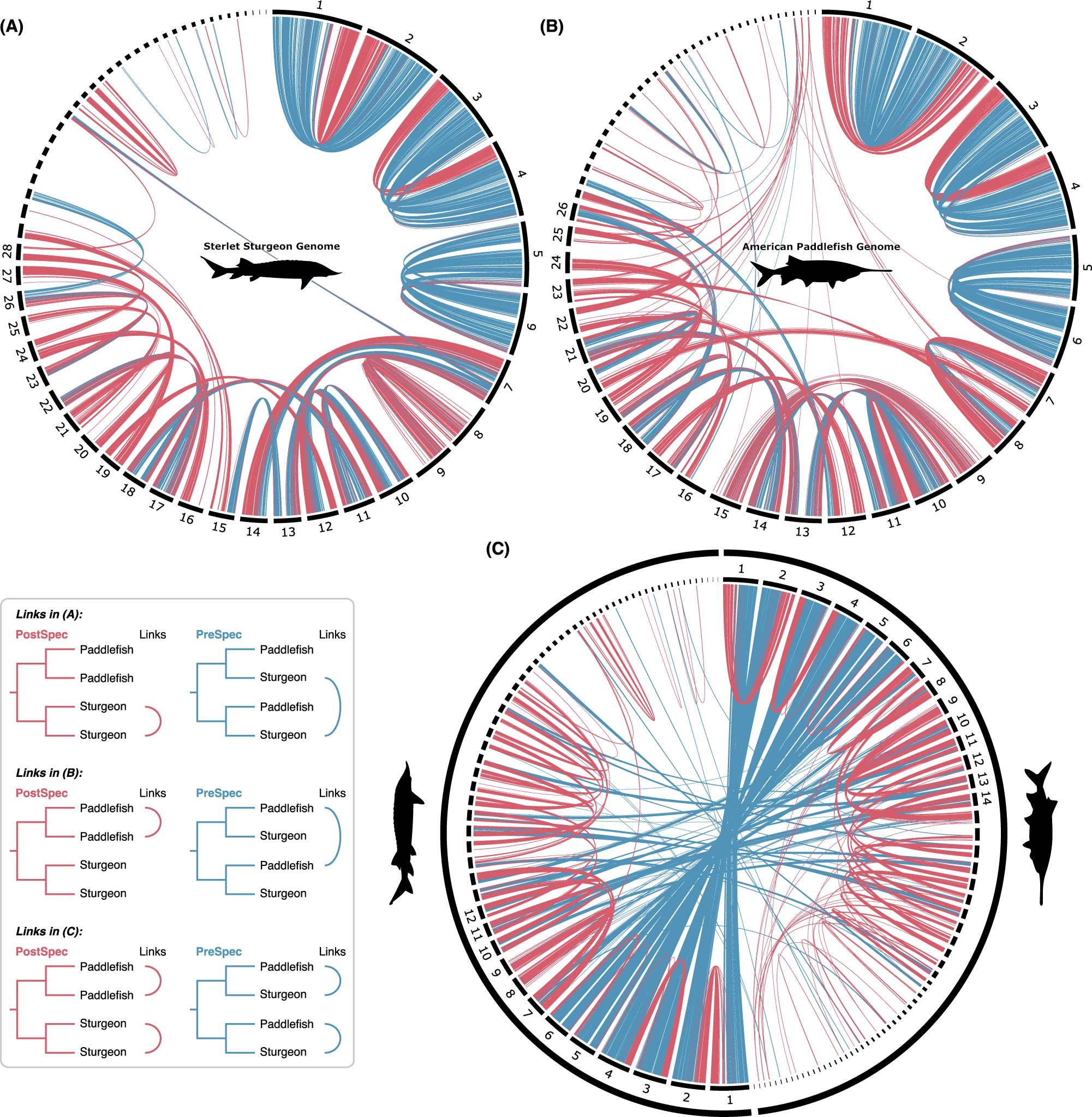 Chromosomal location of ohnolog pairs in sturgeon and paddlefish, from Redmond et al. 2023We are actively researching whole genome duplication (WGD) and rediploidisation. WGD is a major evolutionary event in which the entire genome is doubled. WGD provides much raw material for evolution and may have played a role in survival during mass extinction events.
Chromosomal location of ohnolog pairs in sturgeon and paddlefish, from Redmond et al. 2023We are actively researching whole genome duplication (WGD) and rediploidisation. WGD is a major evolutionary event in which the entire genome is doubled. WGD provides much raw material for evolution and may have played a role in survival during mass extinction events.
Rediploidisation is the process of the genome returning to diploid following WGD. This has recently been shown to have been a delayed and asynchronous process following multiple different vertebrate WGDs. Our team recently showed that sturgeon and paddlefish, who were previously thought to have had independent WGDs, shared a WGD in their common ancestor. This shared WGD was masked due to asynchronous rediploidisation, which resulted in many of the duplicate genes being species-specific. Duplicate genes cannot diverge from each other in sequence and function until they have returned to diploid, allowing for evolution of lineage-specific novelties.
WGD events have occurred many times across the tree of life. We are interested in investigating these processes and their impact in vertebrate evolution.
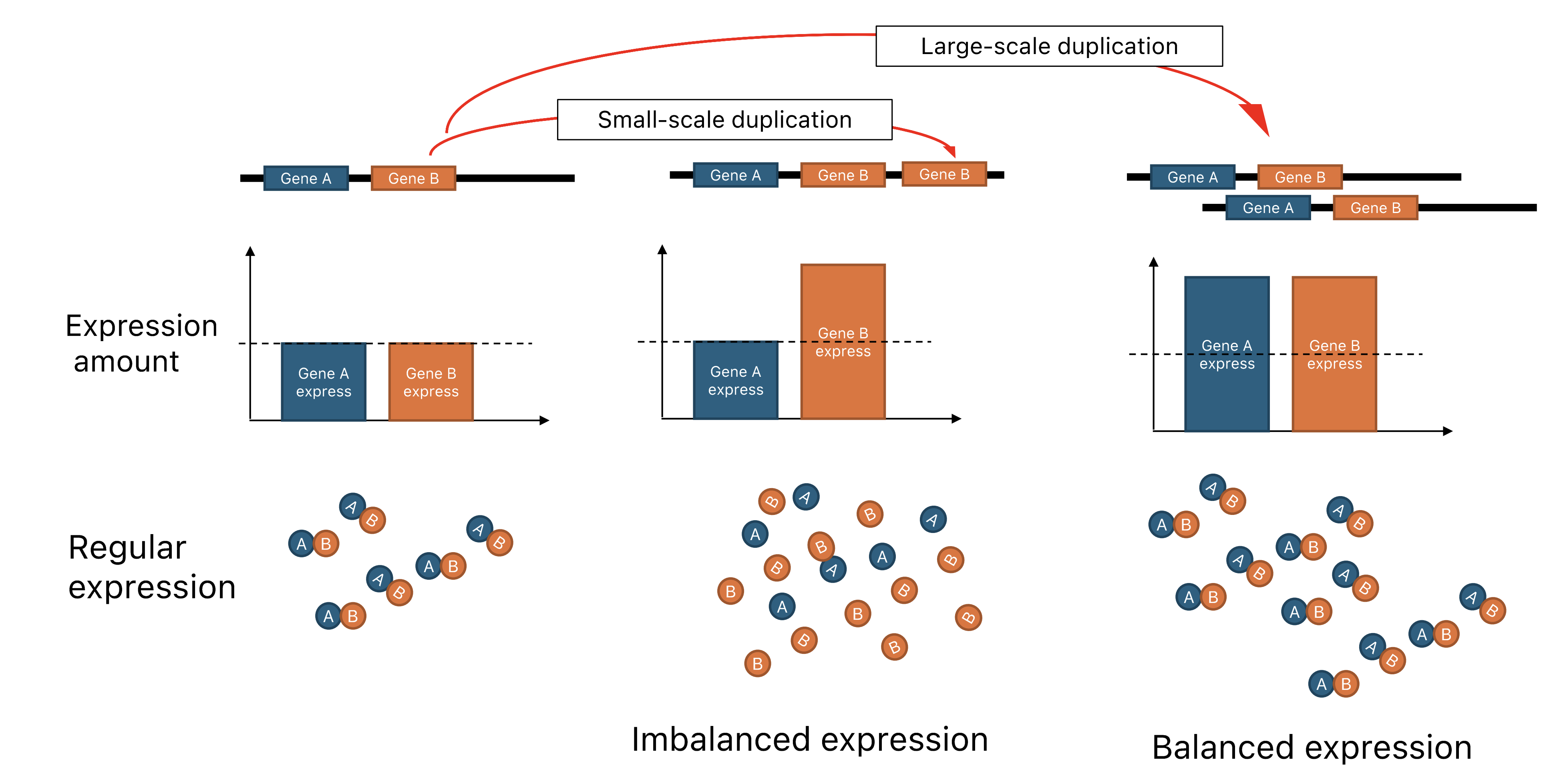 Impact of small-scale duplication vs large-scale duplication on dosage balanceWe are also interested in dosage sensitivity. We have found that dosage sensitivity plays a key role in post-WGD gene evolution. In humans, dosage sensitivity is a major determinant of copy number variant pathogenicity, and dosage constraints also restrict the enrichment of tissue-specific eQTLs in duplicated genes resulting from WGD (ohnologs). A similar phenomenon has been observed in zebrafish in teleost-specific WGD-driven ohnologs. Moreover, in other WGD events across the tree of life, especially in teleost and salmonid WGDs, dosage constraints have led to the accumulation and enrichment of expression divergence associated with subfunctionalization. In summary, our investigation demonstrates the impact of dosage sensitivity on vertebrate evolution after whole genome duplications.
Impact of small-scale duplication vs large-scale duplication on dosage balanceWe are also interested in dosage sensitivity. We have found that dosage sensitivity plays a key role in post-WGD gene evolution. In humans, dosage sensitivity is a major determinant of copy number variant pathogenicity, and dosage constraints also restrict the enrichment of tissue-specific eQTLs in duplicated genes resulting from WGD (ohnologs). A similar phenomenon has been observed in zebrafish in teleost-specific WGD-driven ohnologs. Moreover, in other WGD events across the tree of life, especially in teleost and salmonid WGDs, dosage constraints have led to the accumulation and enrichment of expression divergence associated with subfunctionalization. In summary, our investigation demonstrates the impact of dosage sensitivity on vertebrate evolution after whole genome duplications.
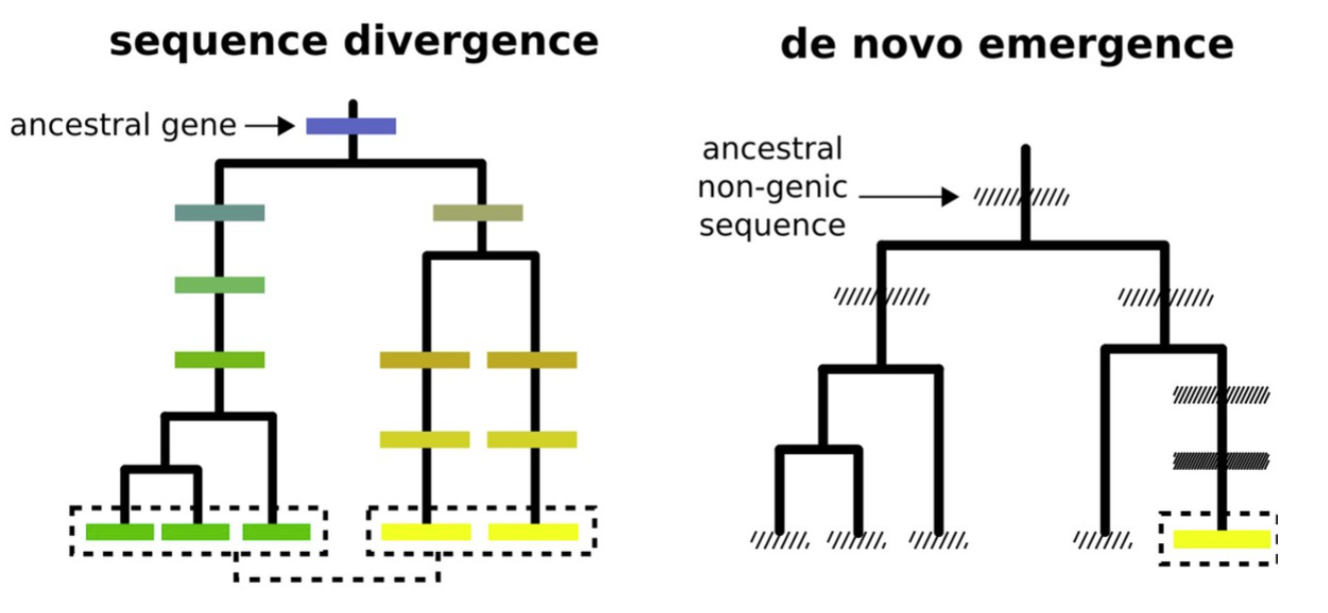 Sequence divergence vs de novo emergence, adapted from Vakirlis et al., 2020As well as studying the mechanisms and impact of gene duplication, we also study de novo gene genesis. This is the process by which genes emerge from previously non-coding regions of the genome (check out McLysaght & Hurst 2016 and McLysaght & Guerzoni 2015 for useful reviews from the lab on this topic). Our team was one of the first to describe the mechanism and rate at which this mechanism of gene evolution occurs, and have carried out work assessing the rate of de novo gene genesis across flies, yeast, and humans. The team was the first to identify the existence of novel human genes. Currently, research in the lab on this area is focusing on new gene evolution in butterflies (Papilionoidea).
Sequence divergence vs de novo emergence, adapted from Vakirlis et al., 2020As well as studying the mechanisms and impact of gene duplication, we also study de novo gene genesis. This is the process by which genes emerge from previously non-coding regions of the genome (check out McLysaght & Hurst 2016 and McLysaght & Guerzoni 2015 for useful reviews from the lab on this topic). Our team was one of the first to describe the mechanism and rate at which this mechanism of gene evolution occurs, and have carried out work assessing the rate of de novo gene genesis across flies, yeast, and humans. The team was the first to identify the existence of novel human genes. Currently, research in the lab on this area is focusing on new gene evolution in butterflies (Papilionoidea).
